Theoretical Overview
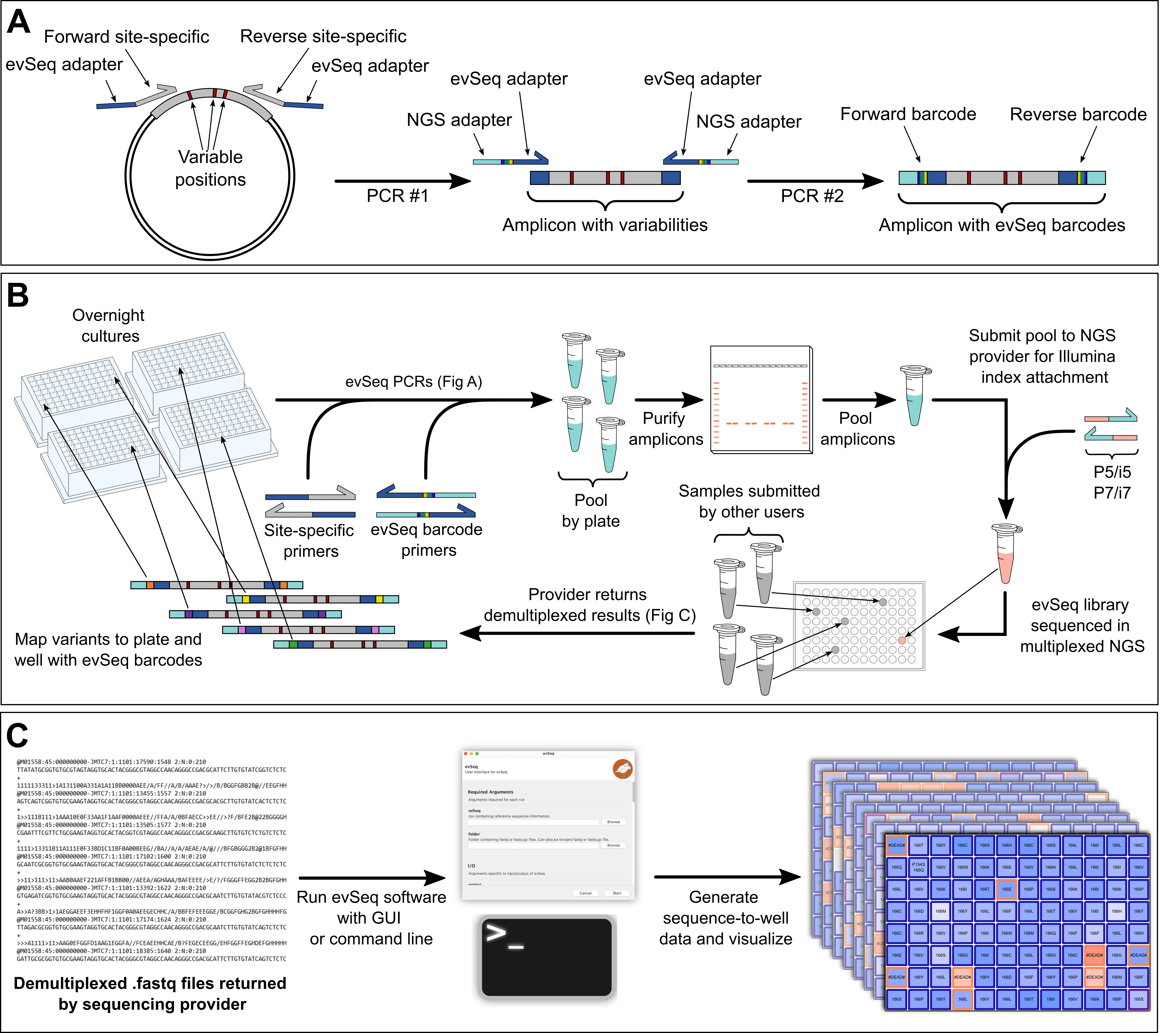
evSeq is an amplicon sequencing procedure designed to make the data in a routine multiplexed NGS experiment go further. This is useful for situations that only call for tens of reads to confidently describe protein variant populations in a single well, where the hundreds of thousands of reads from this type of experiment are unnecessary.
In a typical outsourced multiplexed NGS experiment, samples from multiple researchers are barcoded and pooled. The company running the NGS experiment then sequences the pool and uses the barcodes to de-multiplex the sequencing data; the company then sends each researcher their (and only their) data. For a typical MiSeq NGS run, this spreads the typical 10 - 25 million reads into groups of 100,000 reads – the fewer samples multiplexed, the more reads each researcher gets for their sample.
Meanwhile, in a typical protein engineering experiment, the protein variants that are arrayed into 96-well plates are (ideally) monoclonal, meaning that minimal sequencing information is required to determine the identity of the variant in each well. In a world with perfect sequencing and perfectly monoclonal variants, only a single read would be required. These steps are imperfect though, so more than one read is necessary. However, ~100,000 reads is still very much overkill. Therefore, evSeq was created to further spread these reads over ~100–1000 protein variants, returning enough reads per well for the confident assessment of even polyclonal cultures.
The evSeq protocol was designed to slot into existing workflows and provide hundreds of protein sequences for just cents per variant—or the same price as a handful of Sanger sequencing runs—and it can fully identify each variant in polyclonal cultures. (This is not always possible with Sanger sequencing, which just gives bulk information on the population and not individual DNA molecules.) See the page on library preparation for details on how to use evSeq in the laboratory, and the Computation documentation on how to use the data.
Next page: Library Preparation.
Back to the main page.